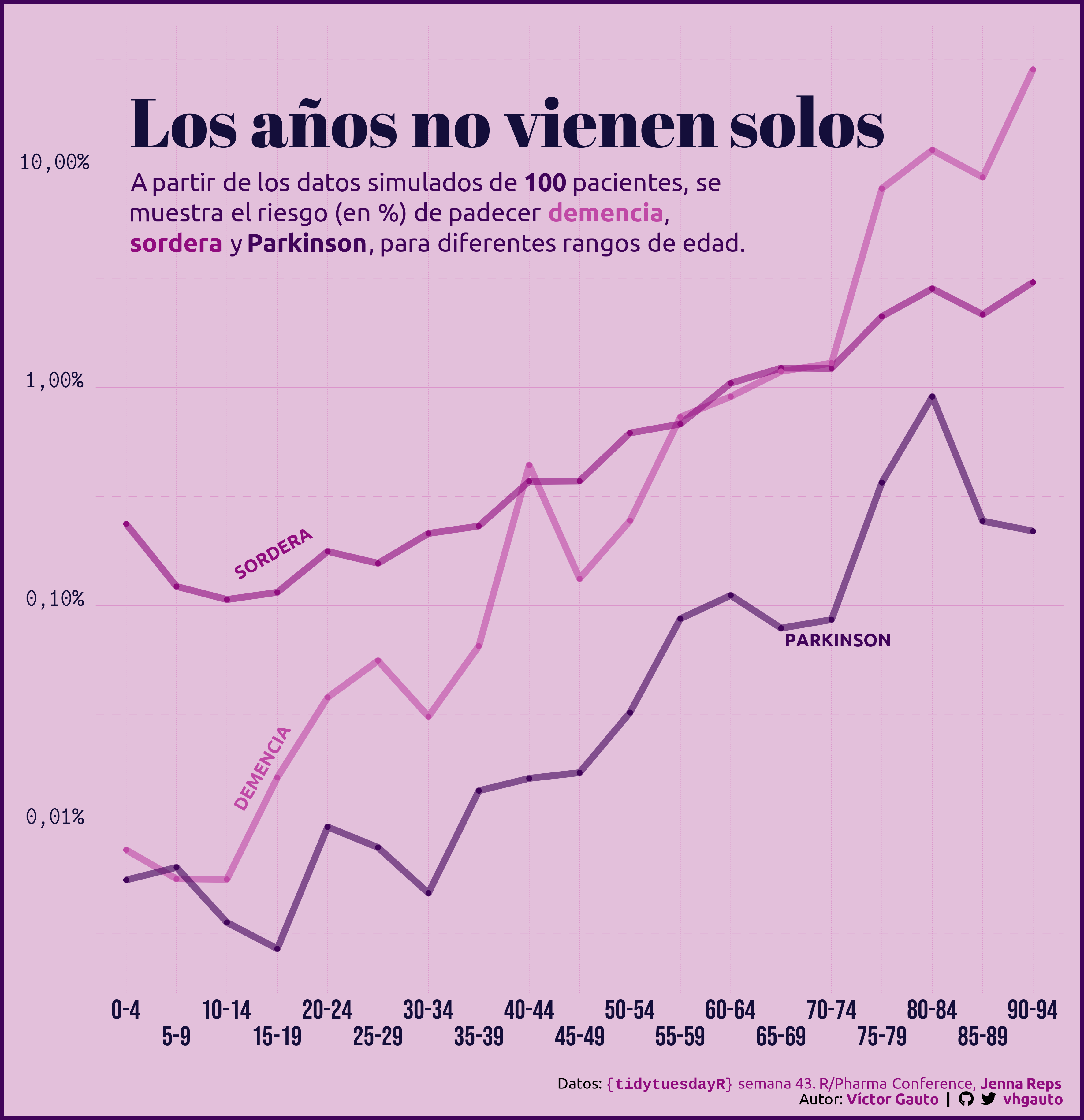
# paquetes ----------------------------------------------------------------
library(tidyverse)
library(showtext)
library(glue)
library(ggtext)
# fuente ------------------------------------------------------------------
# colores
MoMAColors::moma.colors(palette_name = "Flash") |>
as.character()
c1 <- "#E3C0DB"
c2 <- "#41045A"
c3 <- "#900C7E"
c4 <- "#DB95CB"
c5 <- "#140E3A"
c6 <- "#C049A6"
# texto gral
font_add_google(name = "Ubuntu", family = "ubuntu")
# rango de años, eje horizontal
font_add_google(name = "Bebas Neue", family = "bebas")
# porcentajes, eje vertical
font_add_google(name = "Victor Mono", family = "victor", db_cache = FALSE)
# título
font_add_google(name = "Abril Fatface", family = "abril")
# íconos
font_add("fa-brands", "icon/Font Awesome 6 Brands-Regular-400.otf")
font_add("fa-solids", "icon/Font Awesome 6 Free-Solid-900.otf")
font_add("fa-regular", "icon/Font Awesome 6 Free-Regular-400.otf")
showtext_auto()
showtext_opts(dpi = 300)
# caption
fuente <- glue(
"Datos: <span style='color:{c3};'><span style='font-family:mono;'>",
"{{<b>tidytuesdayR</b>}}</span> semana 43. ",
"R/Pharma Conference, ",
"**Jenna Reps**</span>")
autor <- glue("Autor: <span style='color:{c3};'>**Víctor Gauto**</span>")
icon_twitter <- glue("<span style='font-family:fa-brands;'></span>")
icon_github <- glue("<span style='font-family:fa-brands;'></span>")
usuario <- glue("<span style='color:{c3};'>**vhgauto**</span>")
sep <- glue("**|**")
mi_caption <- glue(
"{fuente}<br>{autor} {sep} {icon_github} {icon_twitter} {usuario}")
# datos -------------------------------------------------------------------
browseURL("https://github.com/rfordatascience/tidytuesday/blob/master/data/2023/2023-10-24/readme.md")
patient_risk_profiles <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2023/2023-10-24/patient_risk_profiles.csv')
# me interesa el riesgo de padecer sordera, demencia y Parkinson
# rangos de edad, como factores
rango_orden <- patient_risk_profiles |>
select(contains("age group")) |>
pivot_longer(cols = everything()) |>
distinct(rango = name) |>
mutate(rango = str_remove(rango, "age group: ")) |>
mutate(rango = str_remove_all(rango, " ")) |>
separate_wider_delim(
rango, delim = "-", names = c("i_rango", NA), cols_remove = FALSE) |>
mutate(i_rango = as.numeric(i_rango)) |>
mutate(rango = fct_reorder(rango, i_rango)) |>
arrange(rango)
# enfermedades de interés
enfermedad <- c(
"predicted risk of Sudden Hearing Loss, No congenital anomaly or middle or inner ear conditions",
"predicted risk of Dementia",
"predicted risk of Parkinson's disease, inpatient or with 2nd diagnosis")
# agrupo por enfermedad y rango de edad
d <- patient_risk_profiles |>
select(any_of(enfermedad), contains("age group")) |>
select(oido = 1, demencia = 2, parkinson = 3, everything()) |>
pivot_longer(
cols = -c(oido, demencia, parkinson),
names_to = "rango",
values_to = "es_rango") |>
pivot_longer(
cols = -c(rango, es_rango),
names_to = "enfermedad",
values_to = "frac_enfermedad") |>
mutate(rango = str_remove(rango, "age group: ")) |>
mutate(rango = str_remove_all(rango, " ")) |>
filter(es_rango == 1) |>
mutate(rango = fct(rango, levels = as.character(rango_orden$rango))) |>
arrange(rango) |>
mutate(rango = fct_inorder(rango)) |>
reframe(
n_rango = n(),
frac_enfermedad = mean(frac_enfermedad),
.by = c(rango, enfermedad)) |>
arrange(rango) |>
mutate(rango = fct_inorder(rango))
# figura ------------------------------------------------------------------
# etiquetas de las enfermedades para cada línea
etq_tbl <- tibble(
enf = c("demencia", "sordera", "parkinson") |> str_to_upper(),
color = c(c6, c3, c2),
x = c(3, 3, 14),
y = c(0.012, .15, .078)/100,
angle = c(60, 30, 0)) |>
mutate(label = glue("<b style='color:{color}'>{enf}</b>"))
# título y subtítulo
g_tit <- glue("Los años no vienen solos")
g_sub <- glue(
"A partir de los datos simulados de **100** pacientes, se<br>",
"muestra el riesgo (en %) de padecer <b style='color:{c6}'>demencia</b>, <br>",
"<b style='color:{c3}'>sordera</b> y <b style='color:{c2}'>Parkinson</b>, ",
"para diferentes rangos de edad.")
# figura
g <- ggplot(d, aes(rango, frac_enfermedad, color = enfermedad, group = enfermedad)) +
geom_line(
show.legend = FALSE, linewidth = 2.5, alpha = .6) +
geom_point(
show.legend = FALSE, alpha = 1) +
geom_richtext(
data = etq_tbl, aes(x, y, label = label, angle = angle), show.legend = FALSE,
inherit.aes = FALSE, hjust = 0, vjust = 1, label.color = NA, fill = NA,
family = "ubuntu", size = 5) +
annotate(
geom = "richtext", label = g_tit, color = c5, x = 1, y = .1,
hjust = 0, vjust = 0, family = "abril", size = 19, fill = NA,
label.color = NA) +
annotate(
geom = "richtext", label = g_sub, color = c2, x = 1, y = .1,
hjust = 0, vjust = 1, family = "ubuntu", size = 7, fill = NA,
label.color = NA) +
scale_color_manual(values = c(c6, c3, c2)) +
scale_y_log10(labels = scales::label_percent(
big.mark = "", decimal.mark = ",")) +
coord_cartesian(clip = "off") +
labs(caption = mi_caption) +
guides(
x = guide_axis(n.dodge = 2)) +
theme_void() +
theme(
aspect.ratio = 1,
plot.margin = margin(20, 16.3, 5, 16.3),
plot.background = element_rect(
fill = c1, color = c2, linewidth = 3),
plot.caption = element_markdown(
family = "ubuntu", margin = margin(25, 0, 5, 0), size = 12),
axis.text = element_text(color = c5),
axis.text.x = element_text(
margin = margin(5, 0, 0, 0), size = 21, family = "bebas"),
axis.text.y = element_text(
vjust = 0, margin = margin(0, 5, 0, 0), family = "victor", size = 15),
panel.grid.major.x = element_line(
color = c4, linetype = 3, linewidth = .2),
panel.grid.major.y = element_line(
color = c4, linetype = 1, linewidth = .2),
panel.grid.minor.y = element_line(
color = c4, linetype = "ff", linewidth = .2)
)
# guardo
ggsave(
plot = g,
filename = "2023/semana_43/viz.png",
width = 30,
height = 31,
units = "cm")
# abro
browseURL("2023/semana_43/viz.png")