# paquetes ----------------------------------------------------------------
library(tidyverse)
library(sf)
library(ggrepel)
library(glue)
library(ggtext)
library(showtext)
library(fontawesome)
# fuentes -----------------------------------------------------------------
font_add_google(name = "Poltawski Nowy", family = "poltawski", db_cache = FALSE) # título
font_add_google(name = "Anuphan", family = "anuphan", db_cache = FALSE) # resto del texto
font_add_google(name = "Share Tech Mono", family = "share", db_cache = FALSE) # coordenadas
showtext_auto()
showtext_opts(dpi = 300)
# íconos
font_add("fa-reg", "icon/Font Awesome 5 Free-Regular-400.otf")
font_add("fa-brands", "icon/Font Awesome 5 Brands-Regular-400.otf")
font_add("fa-solid", "icon/Font Awesome 5 Free-Solid-900.otf")
# caption
icon_twitter <- "<span style='font-family:fa-brands;'></span>"
icon_github <- "<span style='font-family:fa-brands;'></span>"
fuente <- "Datos: <span style='color:#a41400;'><span style='font-family:mono;'>{**tidytuesdayR**}</span> semana 16</span>"
autor <- "Autor: <span style='color:#a41400;'>**Víctor Gauto**</span>"
sep <- glue("**|**")
usuario <- glue("<span style='color:#a41400;'>**vhgauto**</span>")
mi_caption <- glue("{fuente} {sep} {autor} {sep} {icon_github} {icon_twitter} {usuario}")
# datos -------------------------------------------------------------------
browseURL("https://github.com/rfordatascience/tidytuesday/blob/master/data/2023/2023-04-18/readme.md")
founder_crops <- readr::read_csv('https://raw.githubusercontent.com/rfordatascience/tidytuesday/master/data/2023/2023-04-18/founder_crops.csv')
# convierto las coordenadas a sf
founder_crops_sf <- st_as_sf(founder_crops,
coords = c("longitude", "latitude"),
crs = st_crs(4326))
# mapa del mundo
world <- rnaturalearth::ne_countries(scale = "medium", returnclass = "sf")
# región de interés, bbox de la base de datos
l <- st_bbox(founder_crops_sf) |>
st_as_sfc() |>
st_as_sf()
# evito errores al recortar el mapa del mundo
sf_use_s2(FALSE)
# recorto el mapa del mundo a la base de datos
world_subset <- st_crop(world, l)
# me interesan las ubicaciones y 'comestibilidad', solo tomo datos únicos
# incluye 'Edible seed/fruit'
sub <- founder_crops_sf |>
drop_na(edibility) |>
distinct(geometry, edibility) |>
mutate(edibility2 = fct_lump_n(f = edibility, n = 3))
# remuevo la categoría que se repite en TODOS los puntos (Edible seed/fruit)
# conservo las restantes, elijo las 3 más frecuentes y lump el resto
sub2 <- founder_crops_sf |>
# remuevo NA
drop_na(edibility) |>
# mantengo ubicaciones únicas
distinct(geometry, edibility) |>
# remuevo la categoría que se repite en todas las ubicaciones
filter(edibility != "Edible seed/fruit") |>
# lump categorías poco frecuentes
mutate(edibility2 = fct_lump_n(
f = edibility,
n = 3,
ties.method = "first",
other_level = "Otros")) |>
# traduzco
mutate(edibility2 = case_match(
edibility2,
"leaves, stems" ~ "Hoja, tallo",
"flowers, stems" ~ "Flor, tallo",
"leaves, root" ~ "Hoja, raíz",
.default = edibility2))
# categoría Otros
sub3 <- sub2 |>
filter(edibility2 == "Otros") |>
mutate(edibility = case_match(
edibility,
"stems" ~ "Tallo",
"rhizomes, stems and leaves," ~ "Rizoma, tallo, hoja",
"bulbs" ~ "Bulbo",
"flowers" ~ "Flor",
"leaves" ~ "Hoja",
.default = edibility))
# figura ------------------------------------------------------------------
# función p/colorear palabras
f_c <- function(x) {
glue("<span style='color:#a41400'>**{x}**</span>")
}
# figura
g1 <- sub2 |>
ggplot() +
# mundo
geom_sf(data = world_subset, fill = "grey90", color = "grey20",
linewidth = .2, linetype = 2) +
# todos los puntos
geom_sf(data = sub |> select(-edibility2),
color = "#007e2e", alpha = 1, size = 1) +
# puntos de las facetas
geom_sf(alpha = .8, color = "#a41400", size = 3, show.legend = TRUE) +
# otros
geom_label_repel(
data = sub3,
aes(label = edibility, geometry = geometry),
color = "#59386c",
label.size = 0,
label.padding = unit(.1, "line"),
fill = alpha("white", .75),
stat = "sf_coordinates",
force = 7,
size = 4.25,
family = "anuphan",
max.overlaps = 20,
min.segment.length = 0) +
# manual
coord_sf(expand = FALSE, clip = "off") +
labs(x = NULL, y = NULL,
title = "Dieta neolítica",
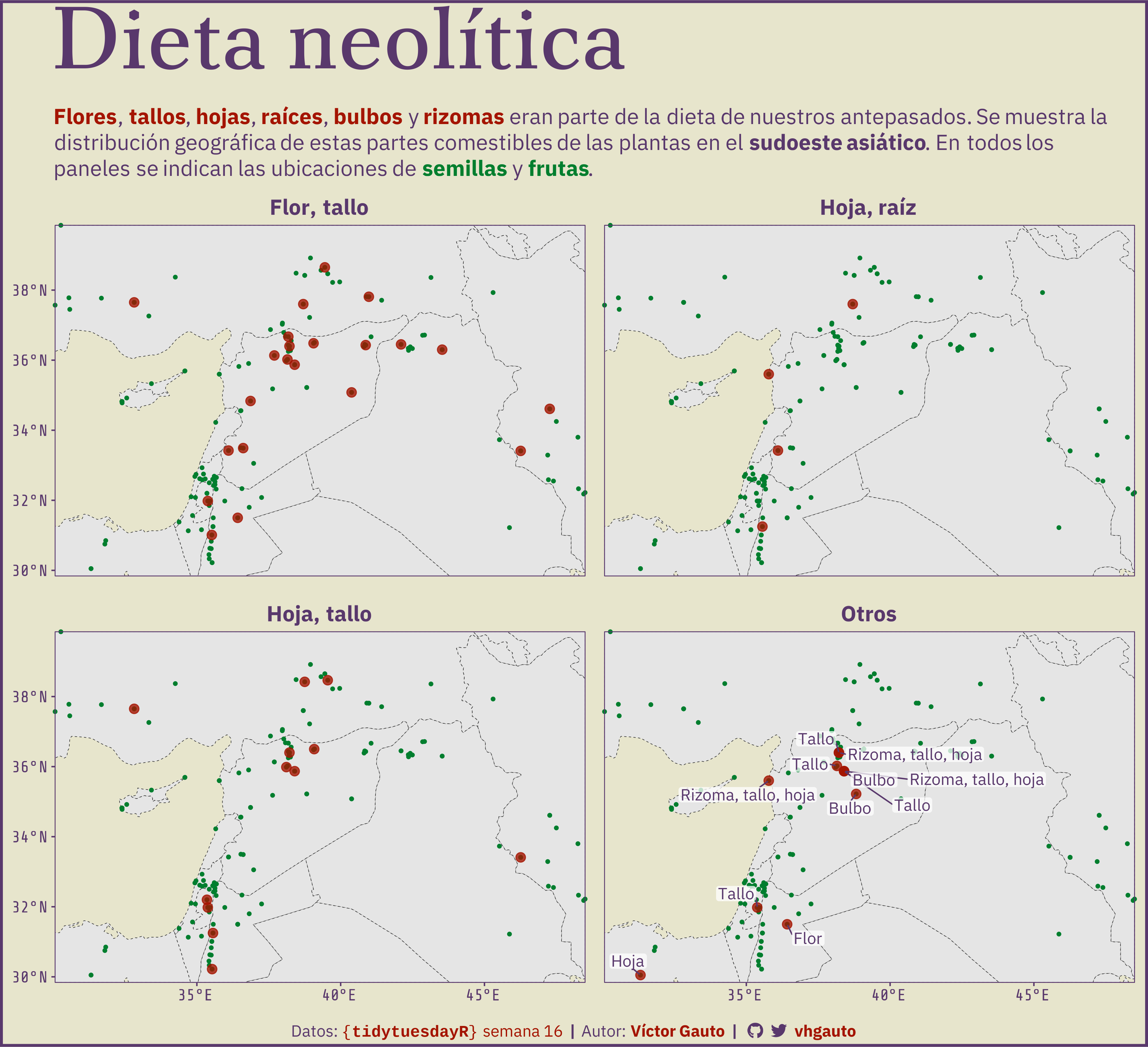
subtitle = glue(
" {f_c('Flores')}, {f_c('tallos')}, {f_c('hojas')}, {f_c('raíces')},
{f_c('bulbos')} y {f_c('rizomas')} eran parte de la dieta de nuestros
antepasados. Se muestra la distribución geográfica de estas partes
comestibles de las plantas en el **sudoeste asiático**. En todos los
paneles se indican las ubicaciones de
<span style='color:#007e2e'>**semillas**</span> y
<span style='color:#007e2e'>**frutas**</span>."),
caption = mi_caption) +
# faceta
facet_wrap(~ edibility2, ncol = 2, nrow = 2) +
# tema
theme_minimal() +
theme(
plot.background = element_rect(
fill = "#e7e5cc", color = "#59386c", linewidth = 2),
plot.title.position = "panel",
plot.title = element_markdown(
size = 65, family = "poltawski", color = "#59386c"),
plot.subtitle = element_textbox_simple(
size = 16, family = "anuphan", color = "#59386c",
margin = margin(10, 0, 10, 0)),
plot.caption = element_markdown(
size = 12, hjust = .46, family = "anuphan", margin = margin(15, 0, 5, 0),
color = "#59386c"),
plot.margin = margin(5, 10, 0, 10),
strip.text = element_markdown(
family = "anuphan", size = 16, color = "#59386c", face = "bold"),
axis.text = element_markdown(family = "share", size = 12, color = "#59386c"),
axis.ticks = element_line(color = "#59386c"),
panel.grid = element_blank(),
panel.ontop = TRUE,
panel.background = element_rect(fill = NA, color = "#59386c", linewidth = .3),
panel.spacing.x = unit(1, "line"),
panel.spacing.y = unit(1.25, "line"))
# guardo
ggsave(
plot = g1,
filename = "2023/semana_16/viz.png",
width = 30,
height = 27.37,
units = "cm",
dpi = 300)
# abro
browseURL("2023/semana_16/viz.png")